Select publications:
Nature Chemical Biology, 2024. 20: p. 142–150. – Full text
Molecular mechanism of GPCR spatial organization at the plasma membrane. Gabriele Kockelkoren, Line Lauritsen, Christopher G. Shuttle, Eleftheria Kazepidou, Ivana Vonkova, Yunxiao Zhang, Artù Breuer, Celeste Kennard, Rachel M. Brunetti, Elisa D’Este, Orion D. Weiner, Mark Uline & Dimitrios Stamou
Nature, 2022. 611: p. 827-834 – Full text
Regulation of the mammalian-brain V-ATPase through ultraslow mode-switching. E. Kosmidis, C. G. Shuttle, J. Preobraschenski, M. Ganzella, P. J. Johnson, S. Veshaguri, J. Holmkvist, M. P. Møller, O. Marantos, F. Marcoline, M. Grabe, J. L. Pedersen, R. Jahn and D. Stamou
Nature Chemical Biology, 2017. 13: p. 724-729 – Full text
Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells. K. R. Rosholm, N. Leijnse, A. Mantsiou, V. Tkach, S. L. Pedersen, V. F. Wirth, L. B. Oddershede, K. J. Jensen, K. L. Martinez, N. S. Hatzakis, P. M. Bendix, A. Callan-Jones and D. Stamou
Science, 2016. 351 (6280): p. 1469-1473 – Full text
Direct observation of proton pumping by a eukaryotic P-type ATPase. S. Veshaguri, S. M. Christensen, G. C. Kemmer, M. P. Møller, G. Ghale, C. Lohr, A. L.Christensen, B. H. Justesen, I. L. Jørgensen, J. Schiller, N. S. Hatzakis, M. Grabe, T. G. Pomorski and D. Stamou
Nature Chemical Biology, 2015. 11 (11): p. 822-825 – Full text
Invited Review Commentary for celebrating the 10th anniversary of Nature Chemical Biology Membrane curvature bends the laws of physics and chemistry. L. Iversen, S. Mathiasen, J. B. Larsen and D. Stamou
Nature Chemical Biology, 2015. 11 (3): p. 192-194 (Front cover page) – Full text
Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases. J.B. Larsen, M.B. Jensen, V.K. Bhatia, S.L. Pedersen, T. Bjørnholm, L. Iversen, M. Uline, I. Szleifer, K.J. Jensen, N.S. Hatzakis and D. Stamou
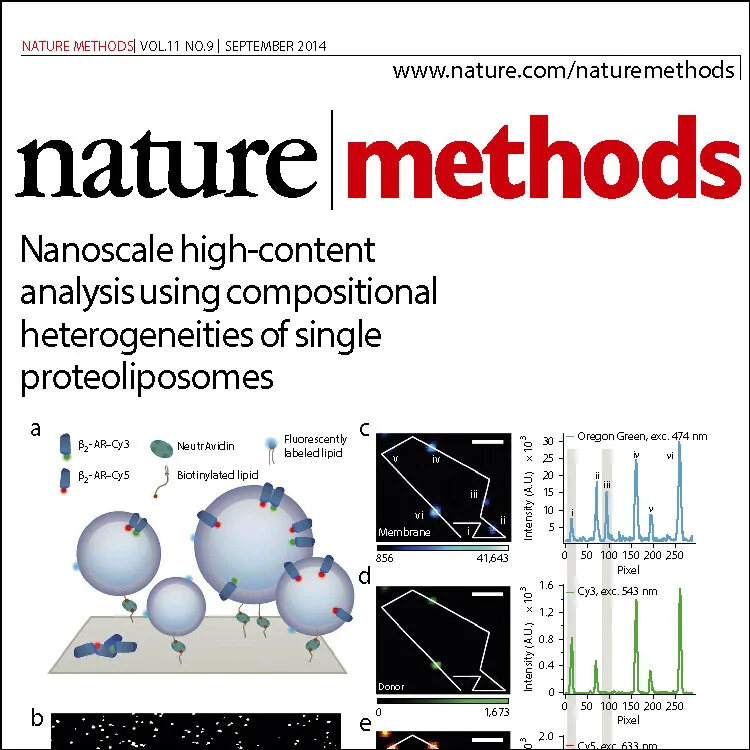
Nature Methods, 2014. 11 (9): p. 931-934 – Full text
Nanoscale high content analysis using compositional heterogeneities of single proteoliposomes. S. Mathiasen, S.M. Christensen,, J.J. Fung, S.G.F. Rasmussen, J.F. Fay, S.K. Joergensen, S. Veshaguri, D.L. Farrens, M. Byrne, B. Kobilka and D. Stamou
Science, 2014. 345 (6192): p. 50-54 – Full text
Single molecule analysis of Ras activation by SOS reveals allosteric regulation via altered fluctuation dynamics. L. Iversen, H.-L. Tu, W.-C. Lin, S. M. Christensen, S. M. Abel, J. Iwig, H.-J. Wu, J. Gureasko, C. Rhodes, R. S. Petit, S. D. Hansen, P. Thill, C.-H. Yu, D. Stamou, A. K. Chakraborty, J. Kuriyan and J. T. Groves.
Nature Nanotechnology, 2012, 7 (1): p. 51–55 – Full text
Mixing sub-attolitre volumes in a quantitative and highly parallel manner with soft matter nanofluidics. S.M. Christensen, P.Y. Bolinger, N.S. Hatzakis, M.W. Mortensen and D. Stamou
Nature Chemical Biology, 2009. 5 (11): p. 835 – Full text
How Curved Membranes Recognize Amphipathic Helices and Protein Anchoring Motifs. N.S. Hatzakis, V.K. Bhatia, J. Larsen, K.L. Madsen, P.Y. Bolinger, A.H. Kunding, J. Castillo, U. Gether, P. Hedegård and D. Stamou
Proceedings of the National Academy of Sciences, 2009. 106 (30): p. 12341 – Full text
Quantification of nano-scale intermembrane contact areas using fluorescence resonance energy transfer. P.M. Bendix, M.S. Pedersen and D. Stamou